Welcome to the RT-Pred Tutorial
This tutorial is composed of 3 parts:
1) Predicting Retention Times;
2) Building a Custom Predictor;
3) Searching RT Data
– Please click the tabs to go to the tutorial of interest. You can access the web server at https://www.rtpred.ca. Enter this web address in your browser to navigate to the website.
This is the tutorial for Predicting Retention Times.
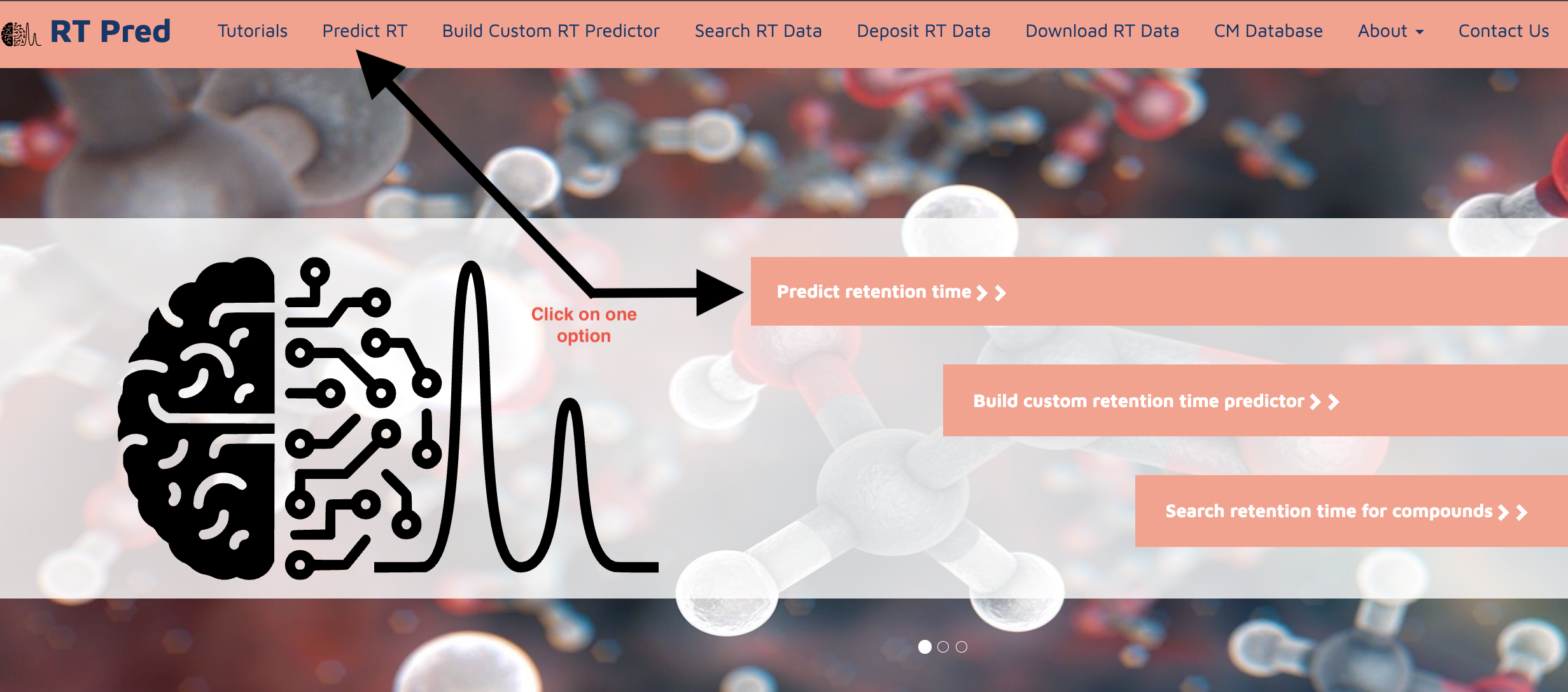
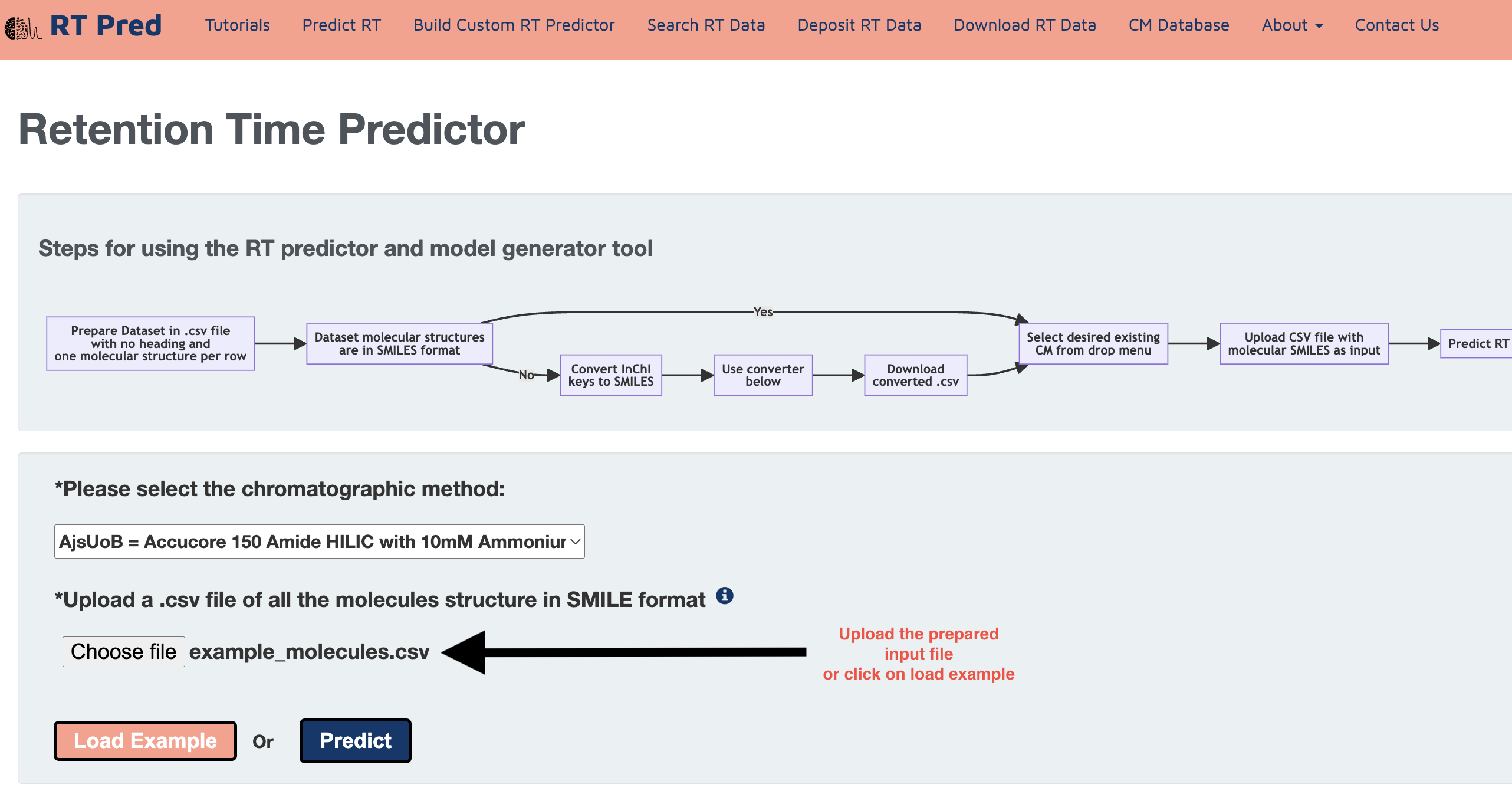
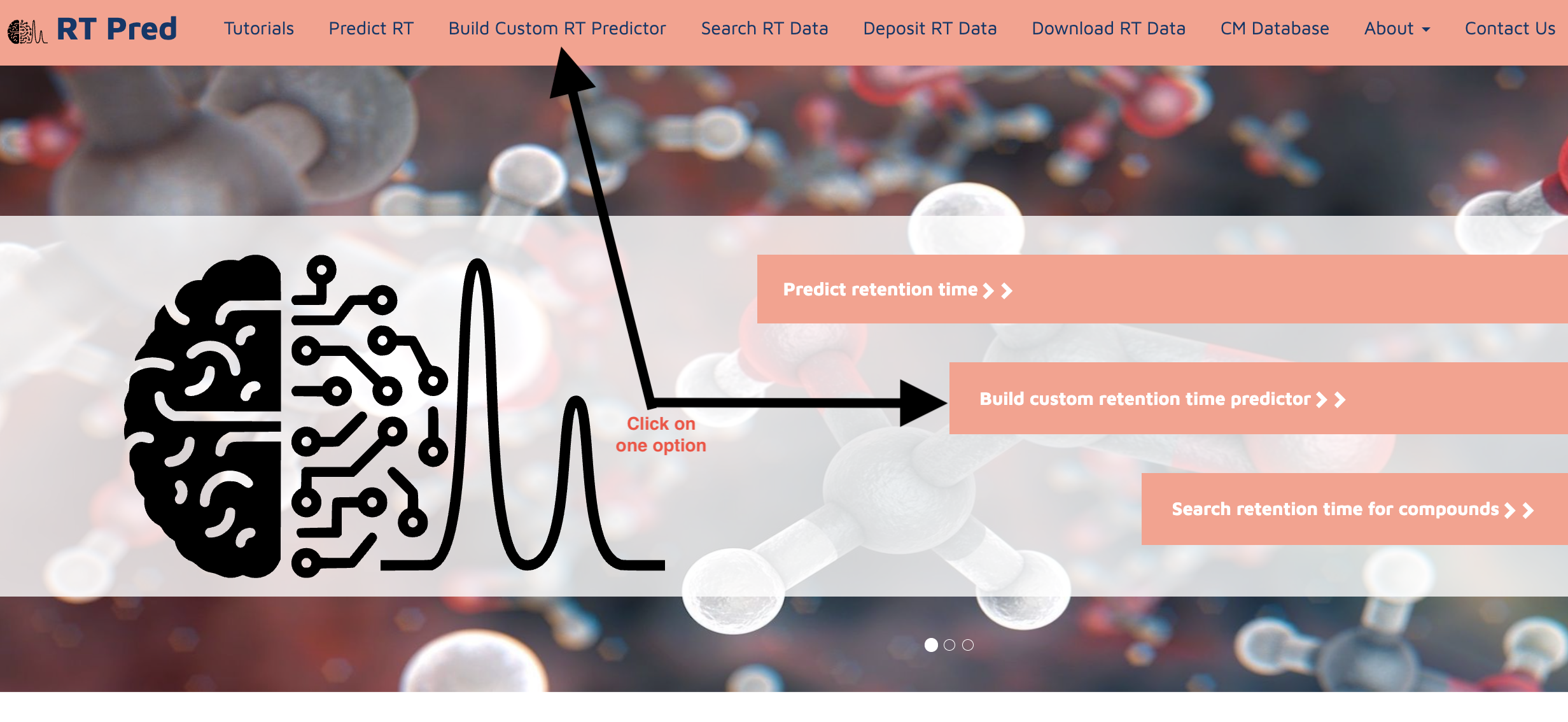
Step 1: Go to the homepage of the server and click on one of the options shown below to go to the predict RT function.
Step 2: Prepare your input file in the following format.
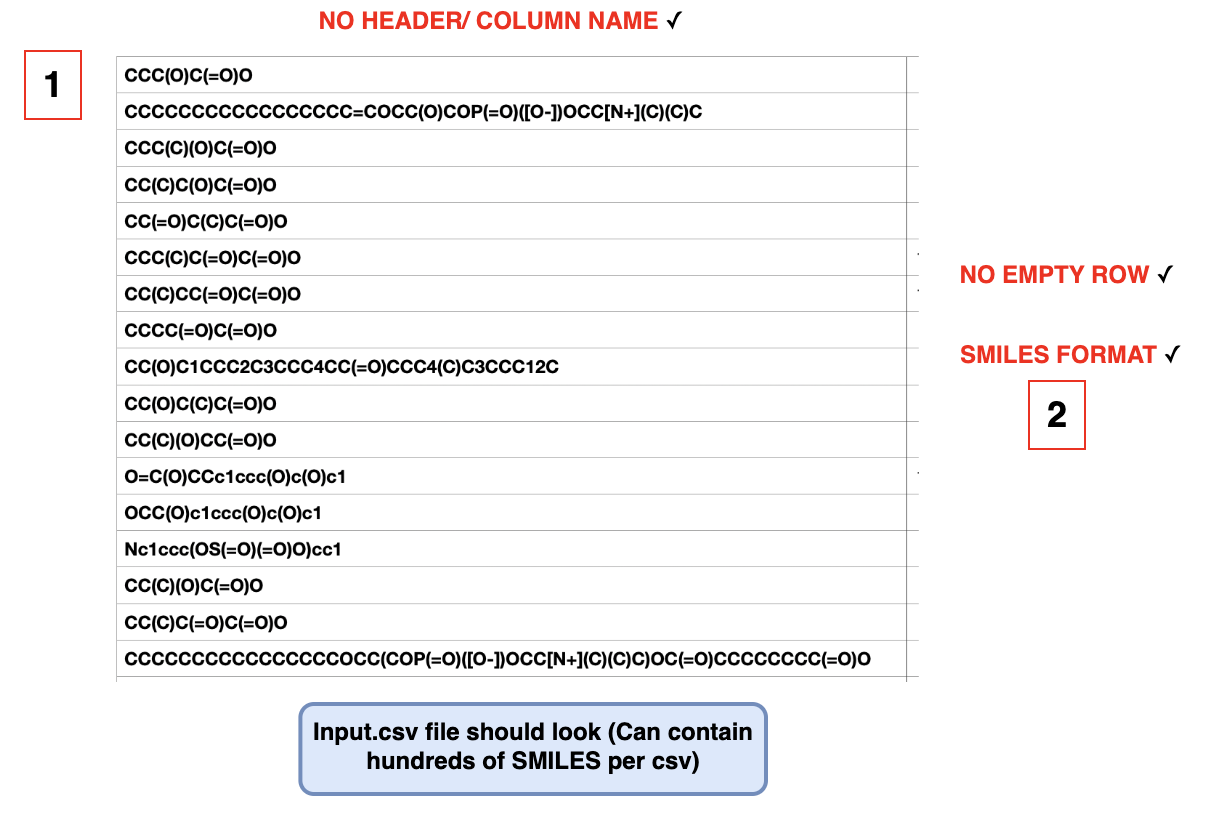
It must be a CSV file with no heading and one molecular structure per row, with no empty rows in between.
Please also ensure that your molecular structures are in SMILES format.
If you have InChI structures, click here to use our InChI to SMILES converter. (Click here)
Step 3: Choose a pretrained Chromatographic Method from the drop down menu.
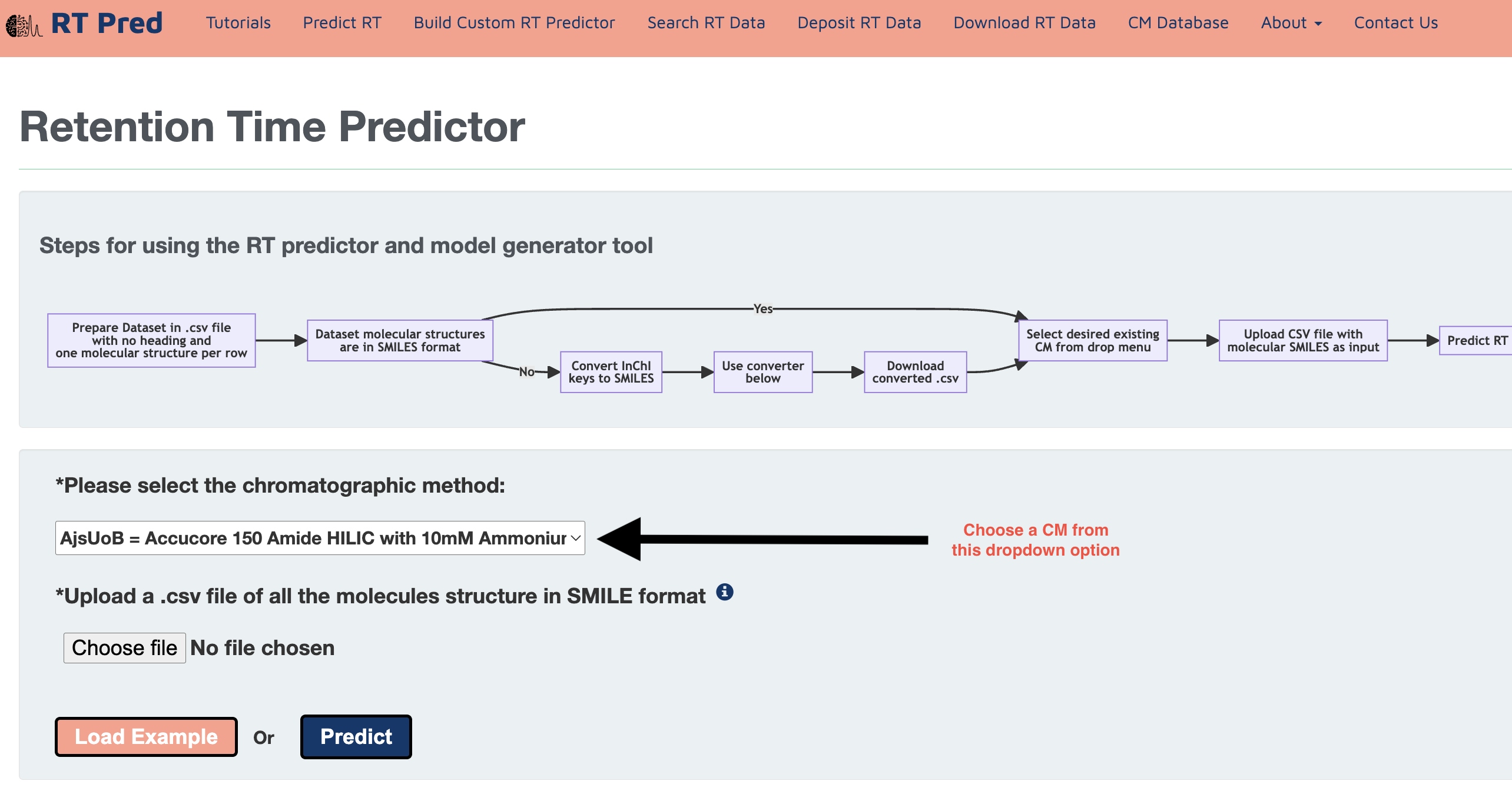
Step 4: Click on the choose file option to upload the prepared input file.
Alternatively, click on the load example button to add a prepared example input.csv file.
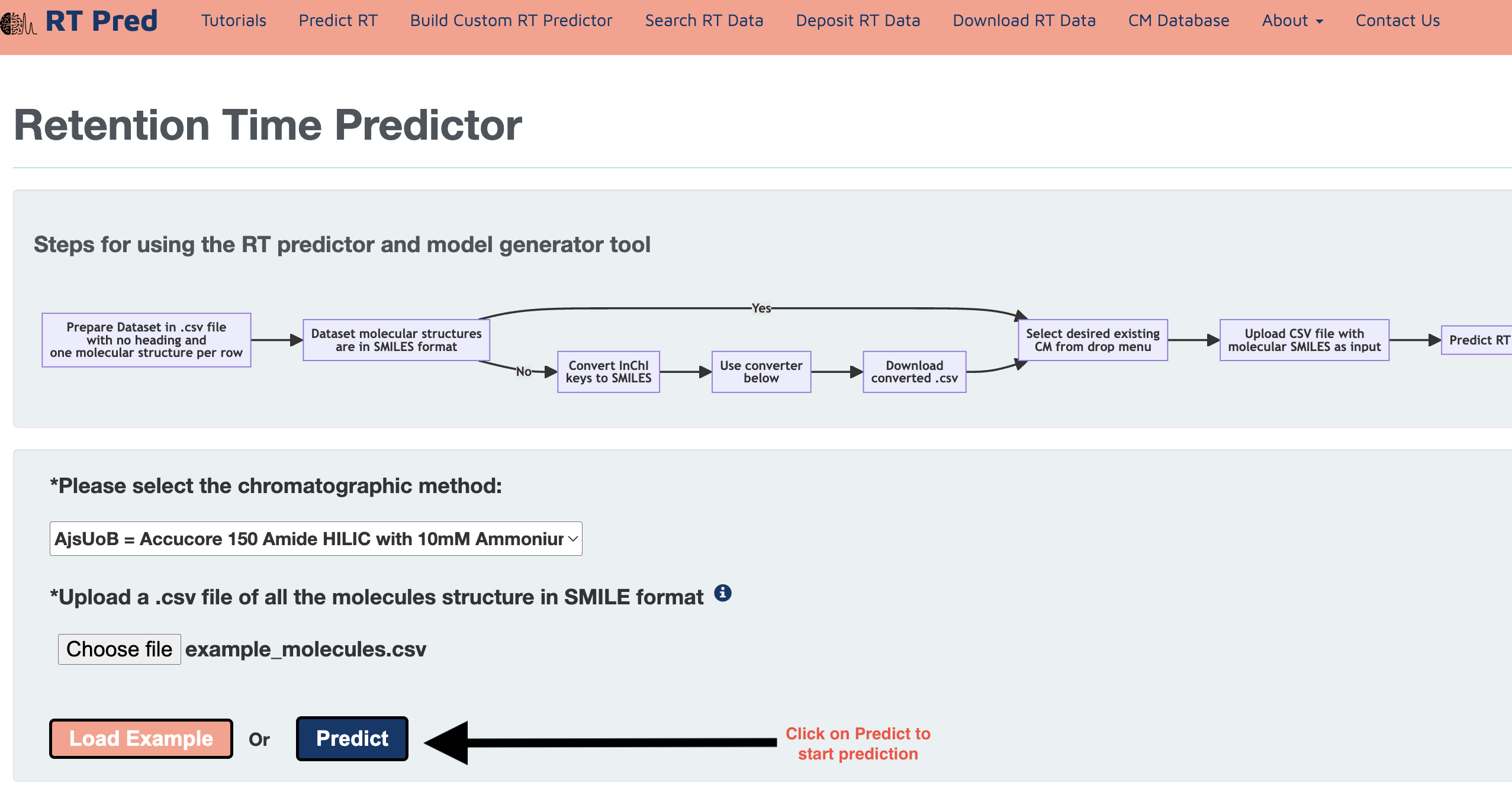
Step 5: Click on the predict button to start prediction.
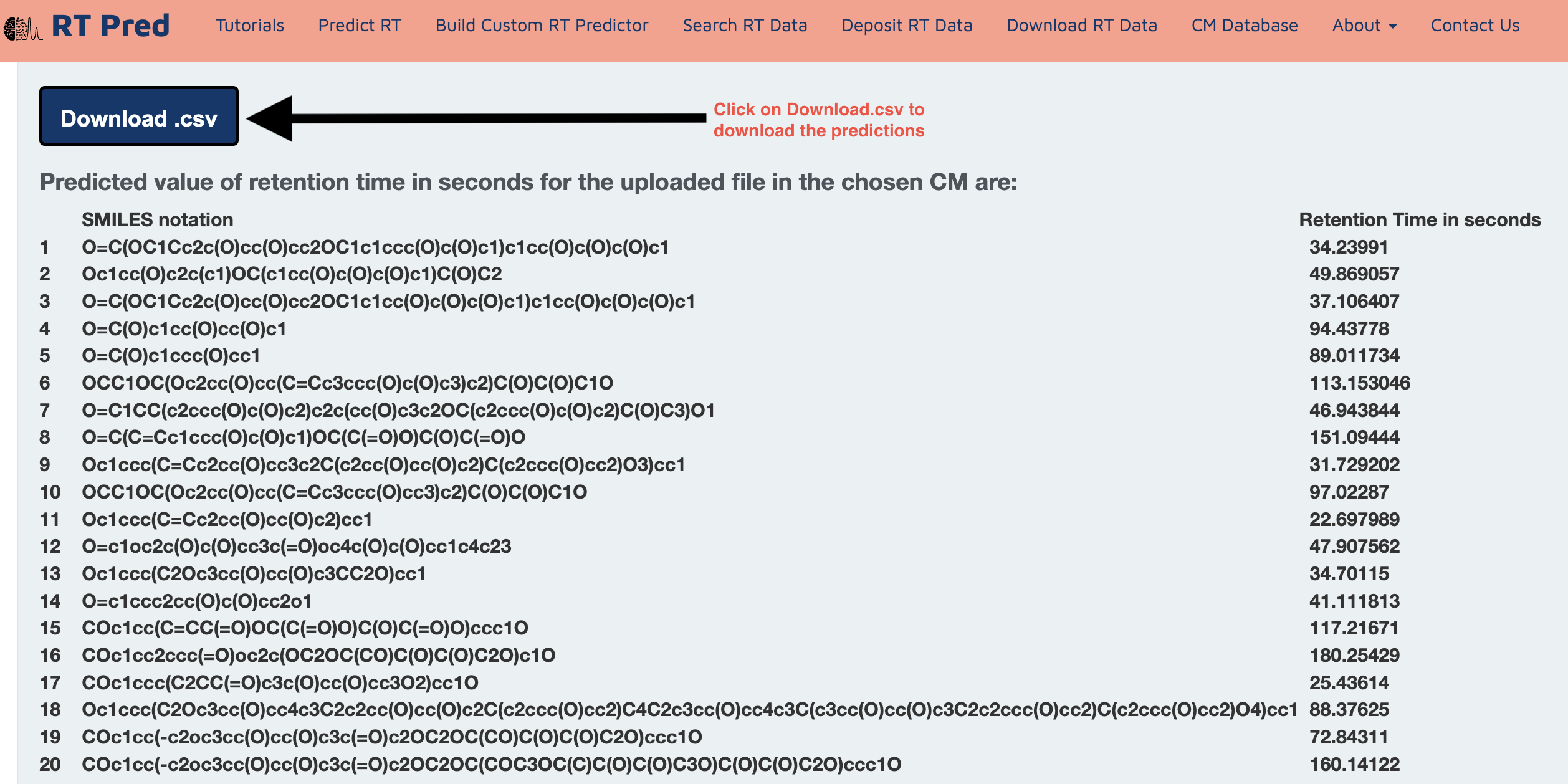
Step 6: Click on the Download.csv button to download a csv file containing your predicted results.
This is the tutorial for Building a Custom Retention Time Predictor.
Step 1: Go to the homepage of the server and click on one of the options shown below to go to the build custom RT model function.
Step 2: Prepare your input file in the following format.

It must be a CSV file with no heading and no empty rows in between.
Please also ensure each row contains one molecular structure in SMILES format in one column and its corresponding retention time in the next column.
If you have InChI structures, click here to use our InChI to SMILES converter. (Click here)
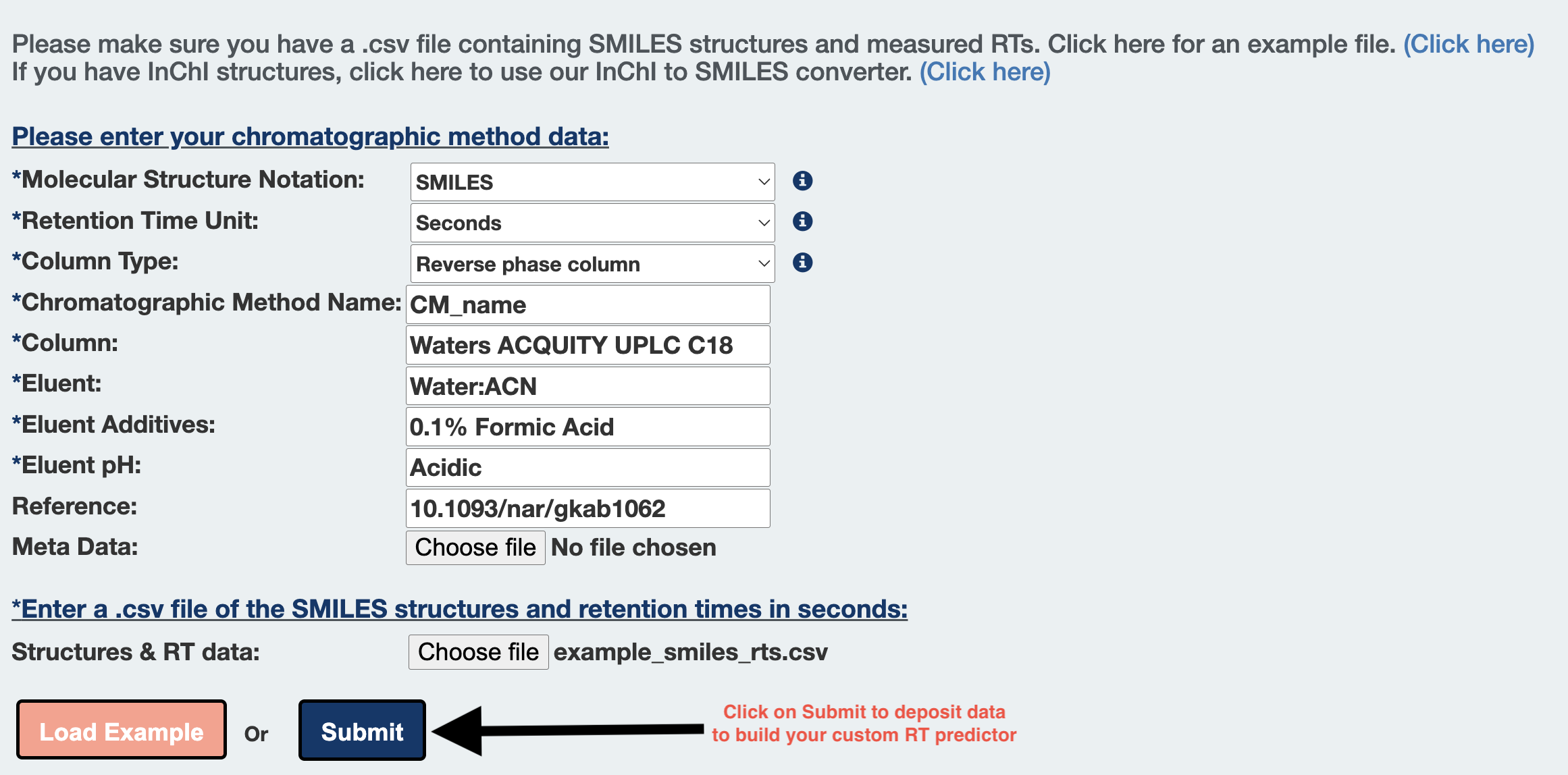
Step 3: Choose options from the dropdowns and fill out the text fields to add details about your chromatographic method.
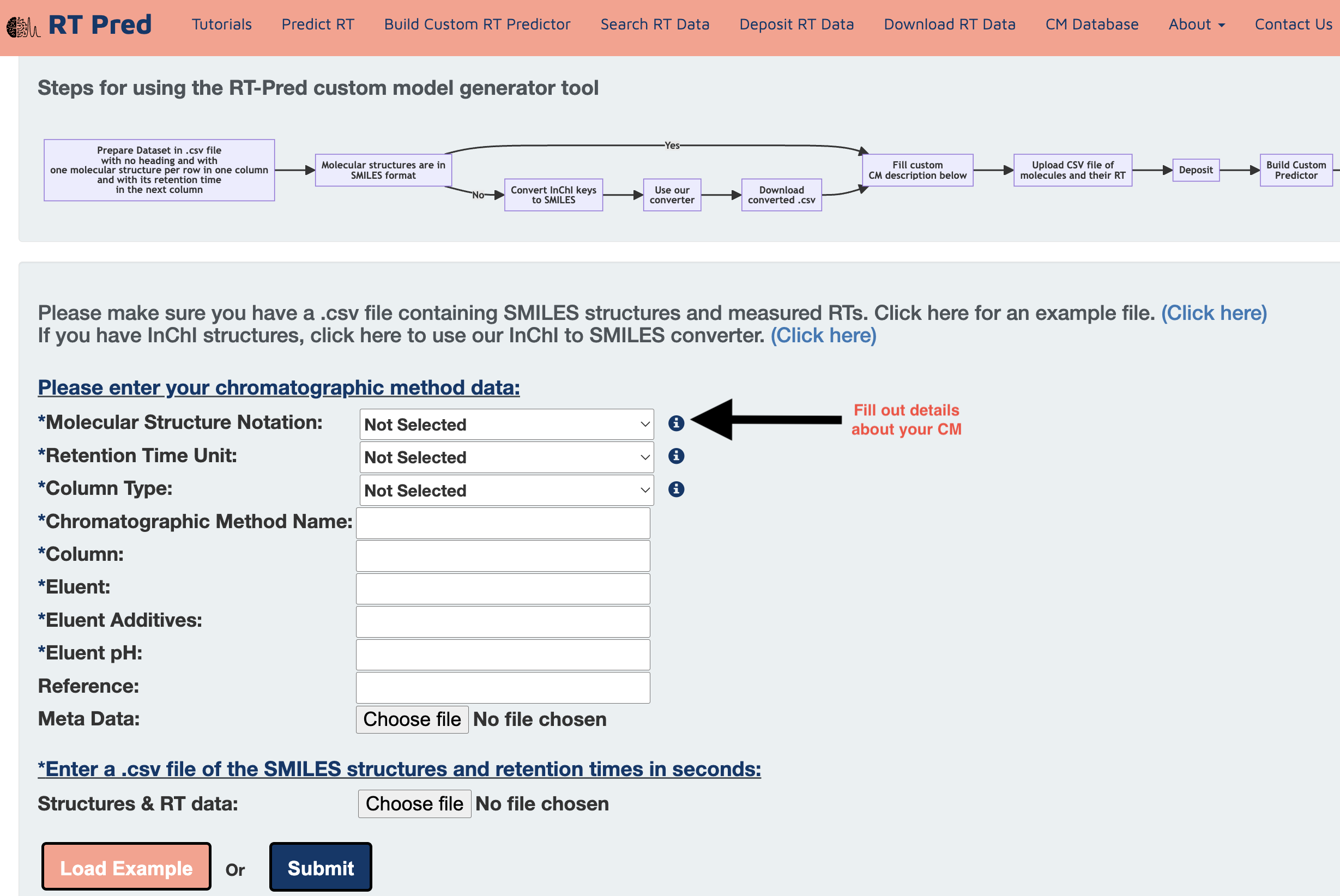
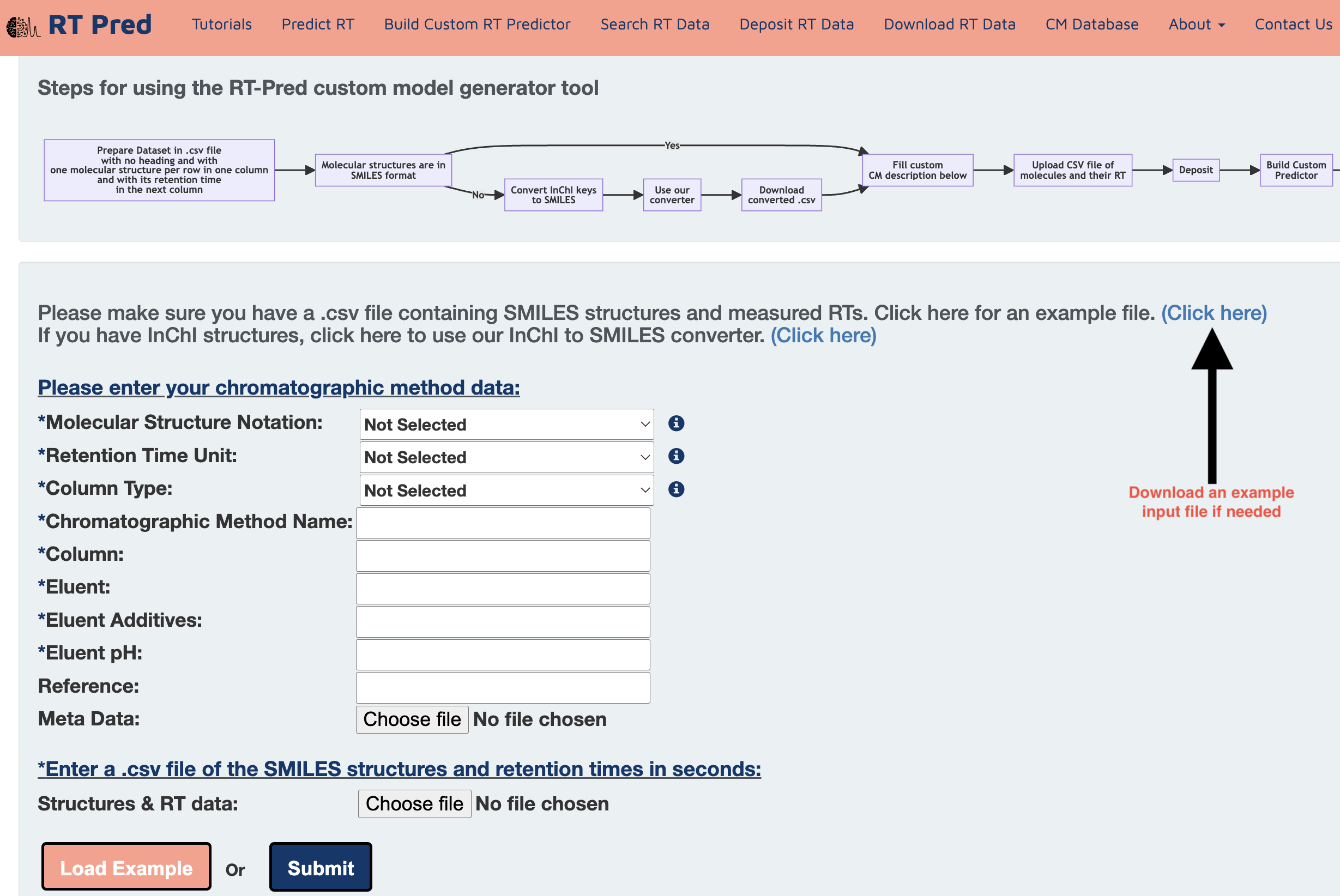
Step 4: Click here to download an example input file containing SMILES structures and measured RTs.
Alternatively, click on the load example button to add a prepared example input.csv file.
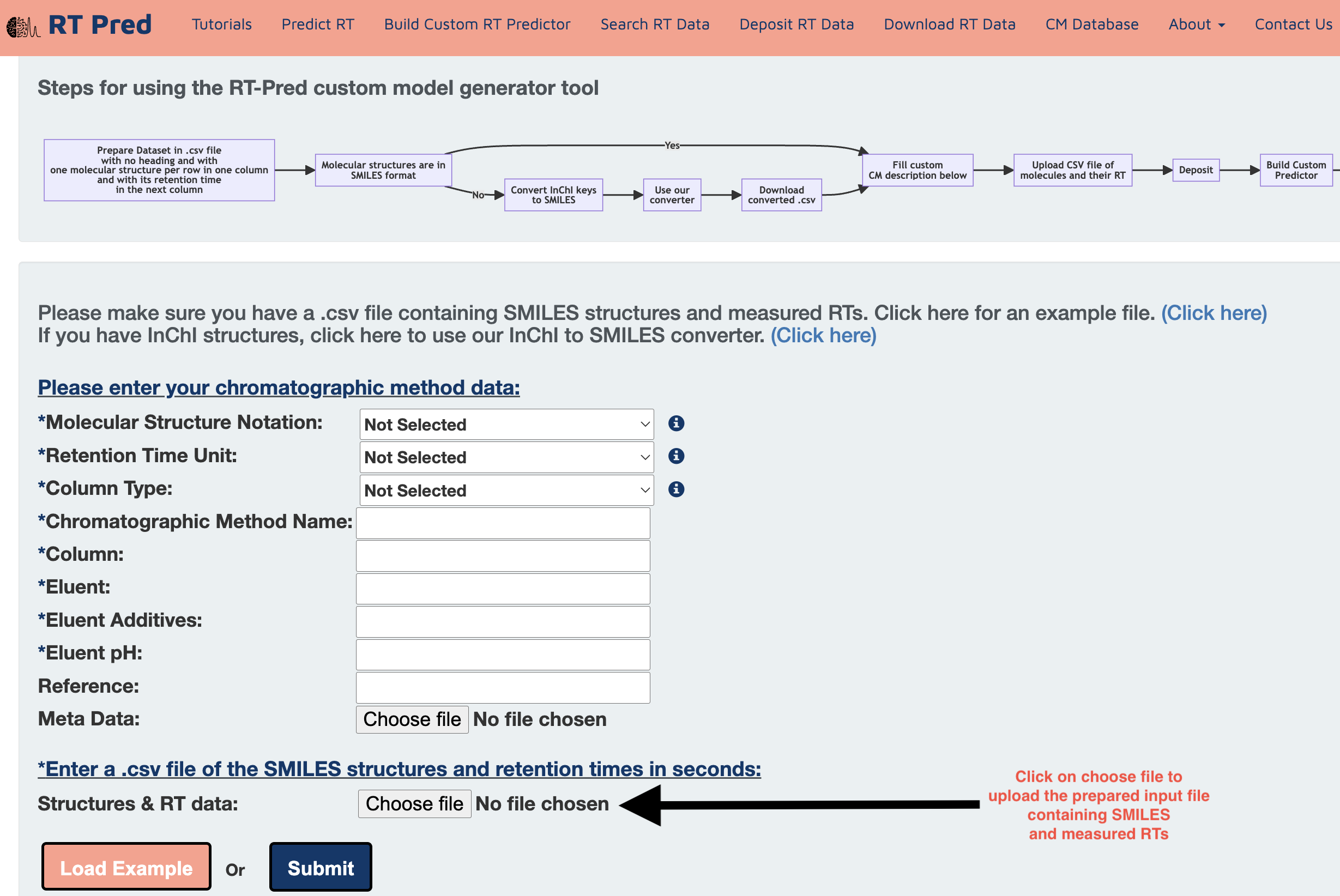
Step 5: Click on choose file to upload your custom CM dataset, formatted according to Step 2.
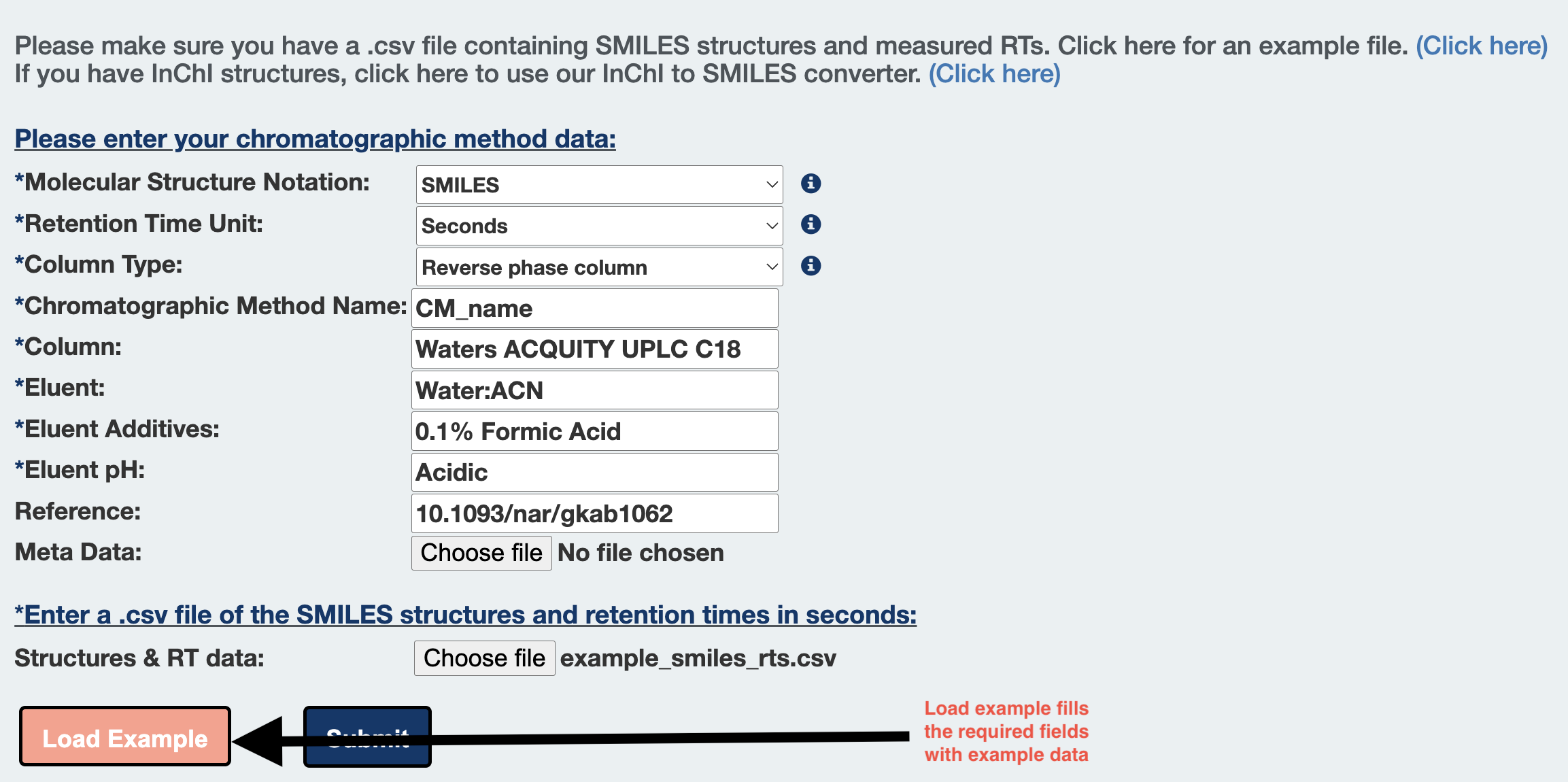
Step 6: Alternatively, click on Load Example button to fill an example CMs description and input file for trying out the server.
Step 7: Click on submit to deposit data to start building your custom RT predictor.
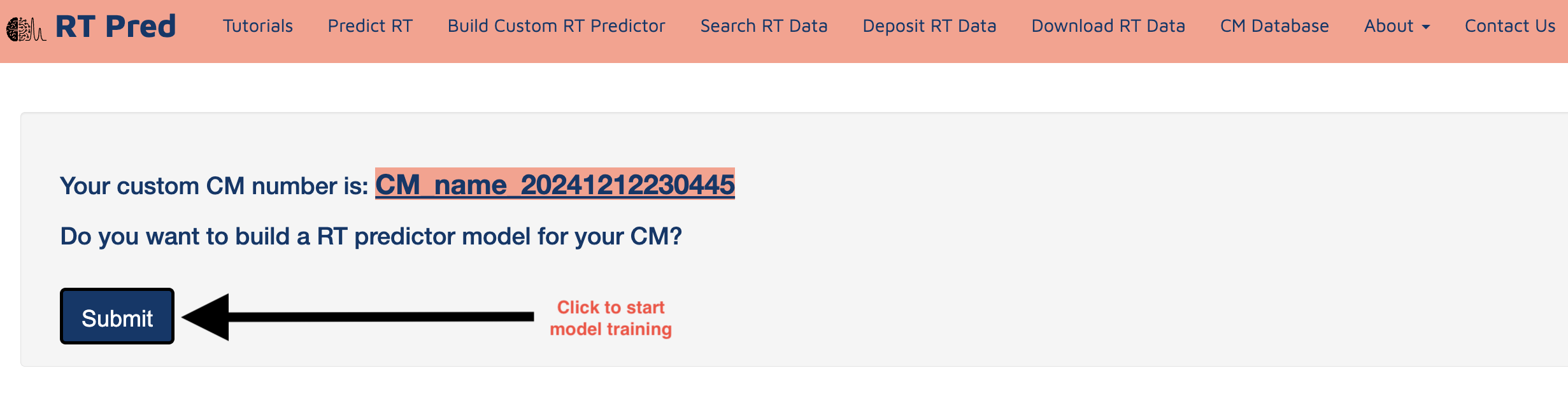
Step 8: Click on submit to start model training for your custom RT predictor.
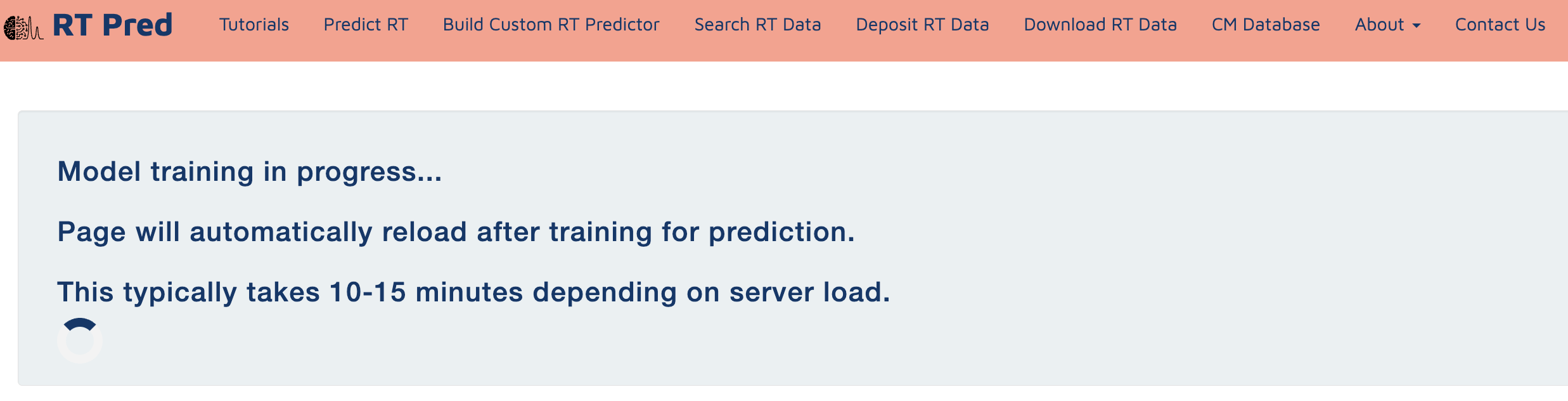
Step 9: Model training in progress.
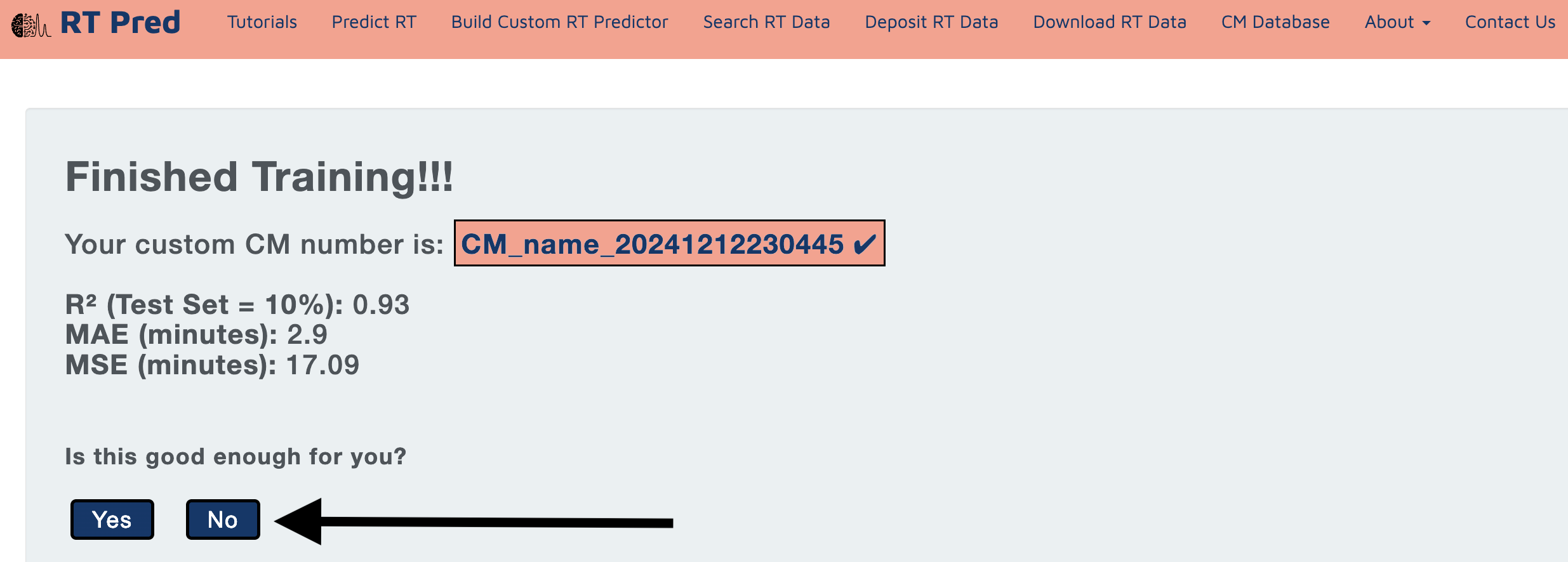
Step 10: Model training finished.
The model uses a 10% held out set from the dataset to test and present the performance statistics.
Click on Yes or No based your performance requirements.
Clicking on Yes will proceed to the next steps for prediction.
Clicking on No will redirect you to the Build Custom RT model page.
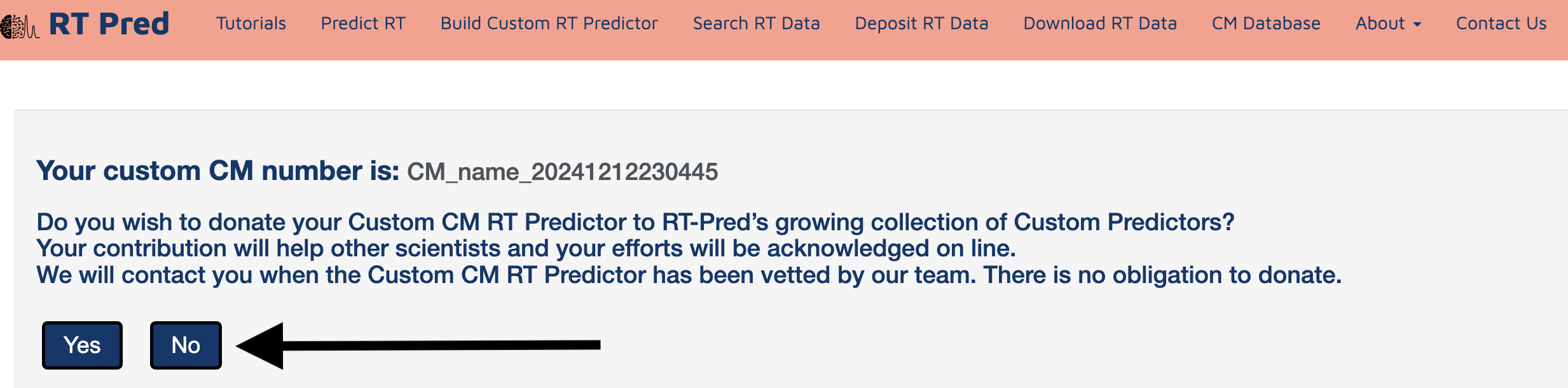
Step 11: This page optionally asks you (with the yes and no option) if you want to donate your model for future research or want it to be deleted from the system.
Clicking either Yes or No, will both redirect you to the prediction page for your custom model.
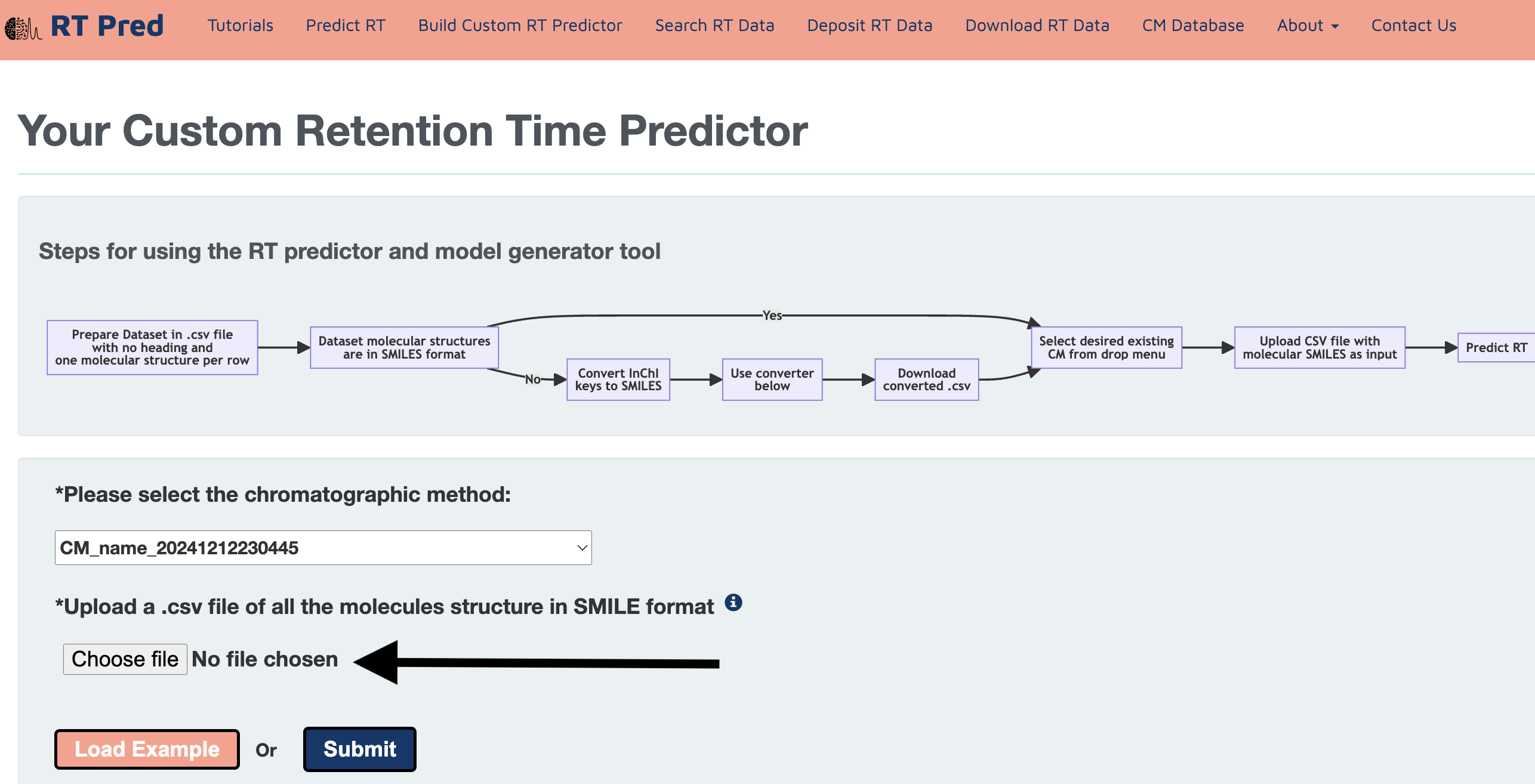
Step 12: Click on choose file to upload a set of SMILES structures for which you want to predict RT in your custom CM.
Alternatively, click on the load example button to add a prepared example input.csv file.
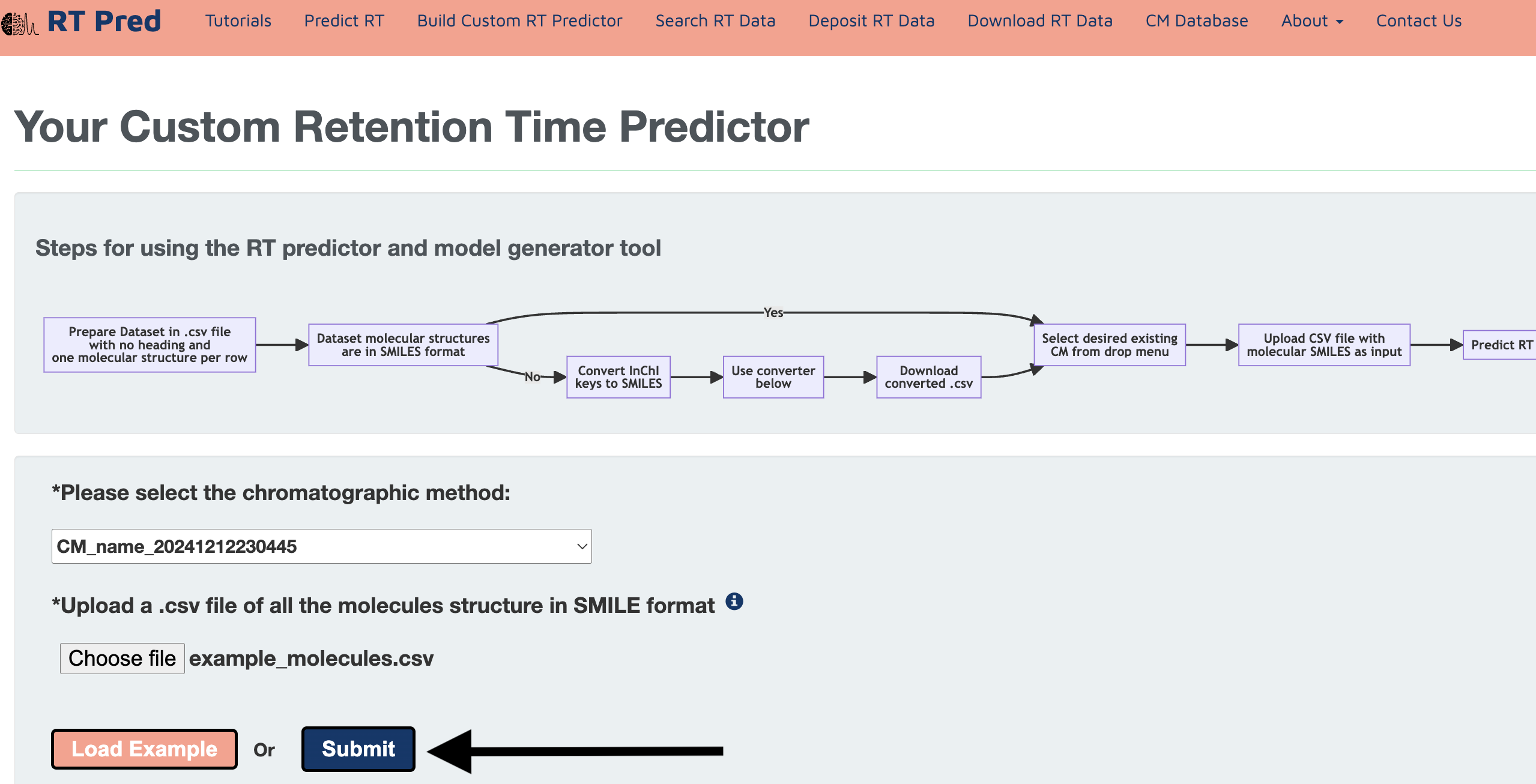
Step 13: Click on the predict button to start prediction.
Step 14: Click on the Download.csv button to download a csv file containing your predicted results.

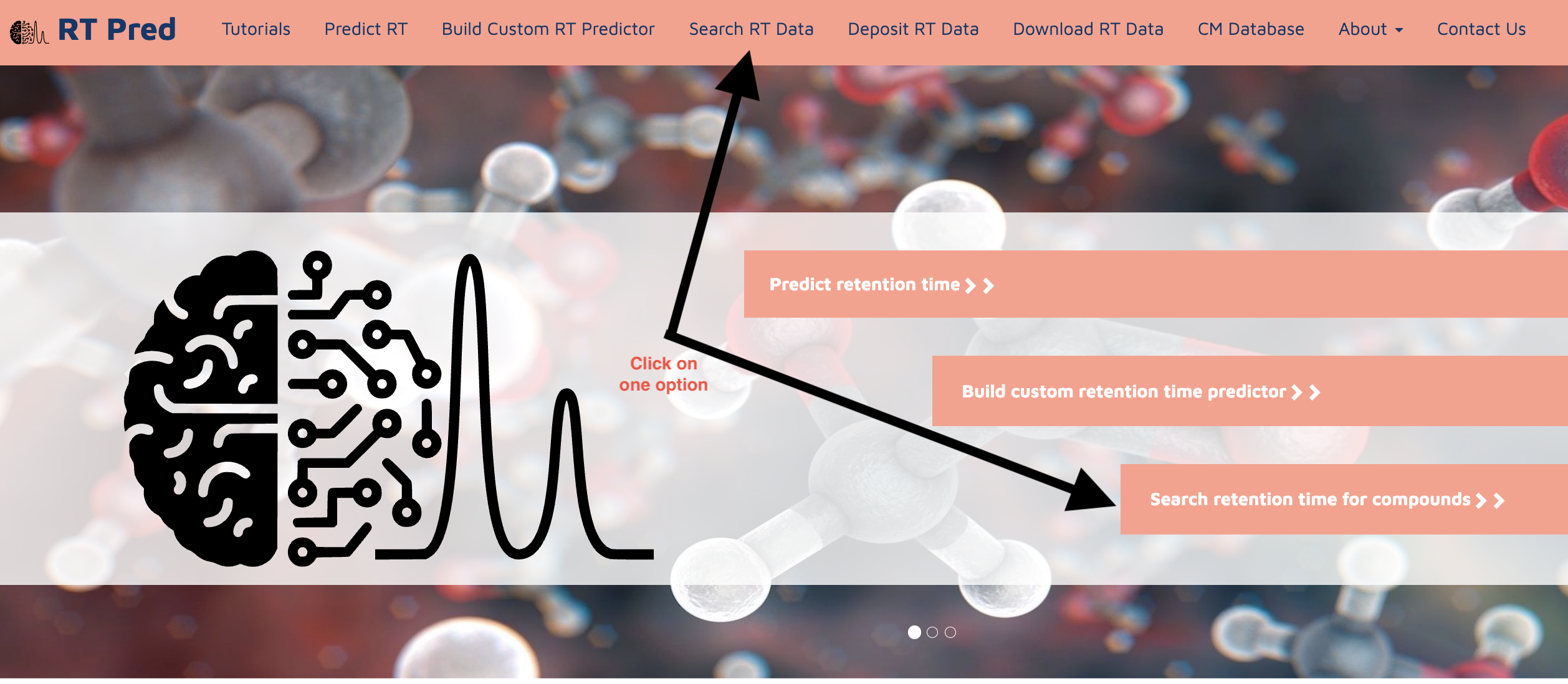
This is the tutorial for Searching RT Data.
Go to the homepage of the server and click on one of the options shown below to go to the search RT function.
Search using Compound to RT
Step 1: Choose a pretrained Chromatographic Method from the drop down menu.
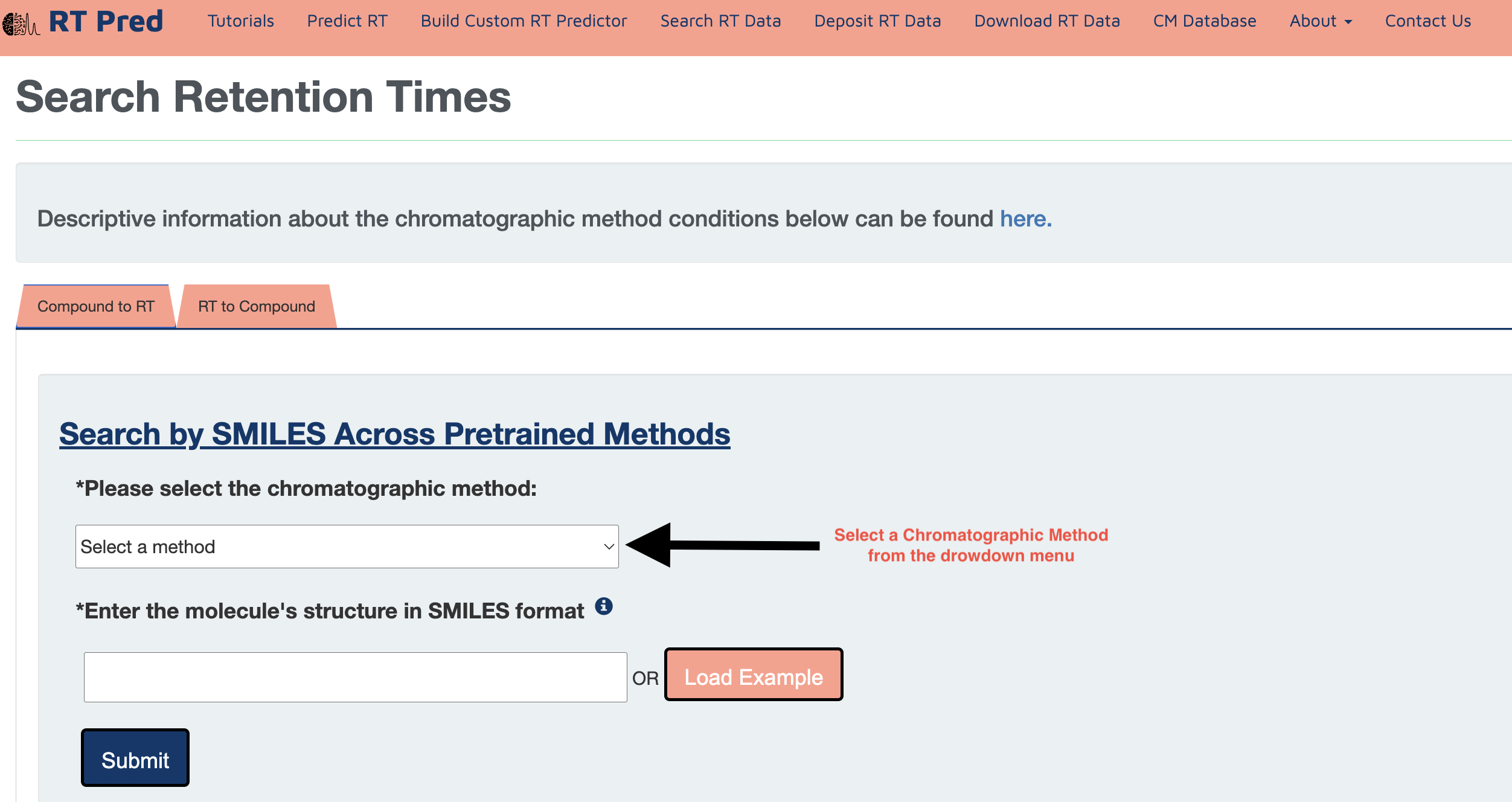
Step 2: Enter a SMILES structure of a compound in the text field.
Alternatively, click on the load example button to add an example SMILES.
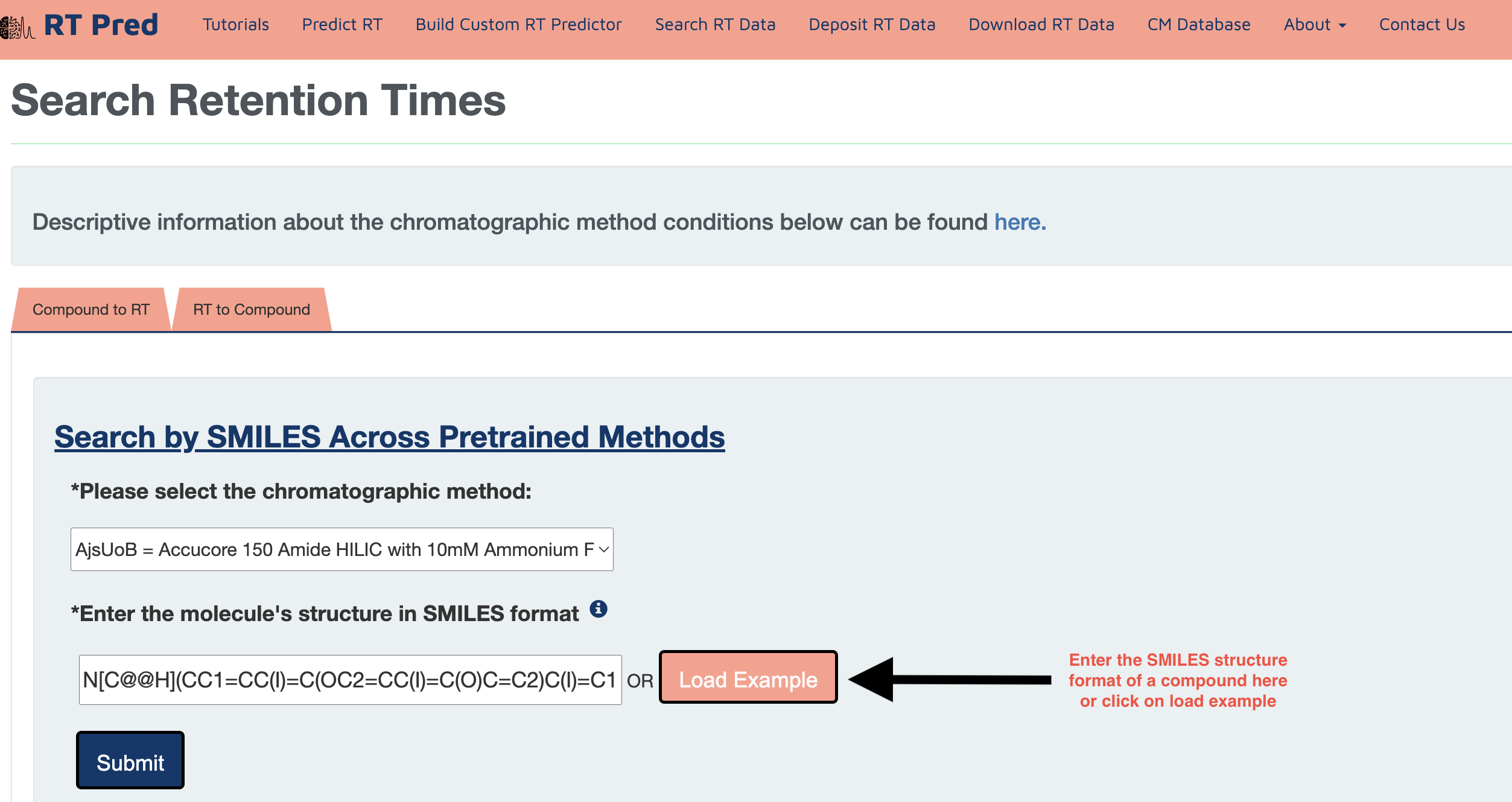
Step 3: Click on the submit button to search for the retention time of the given compound in the selected CM.
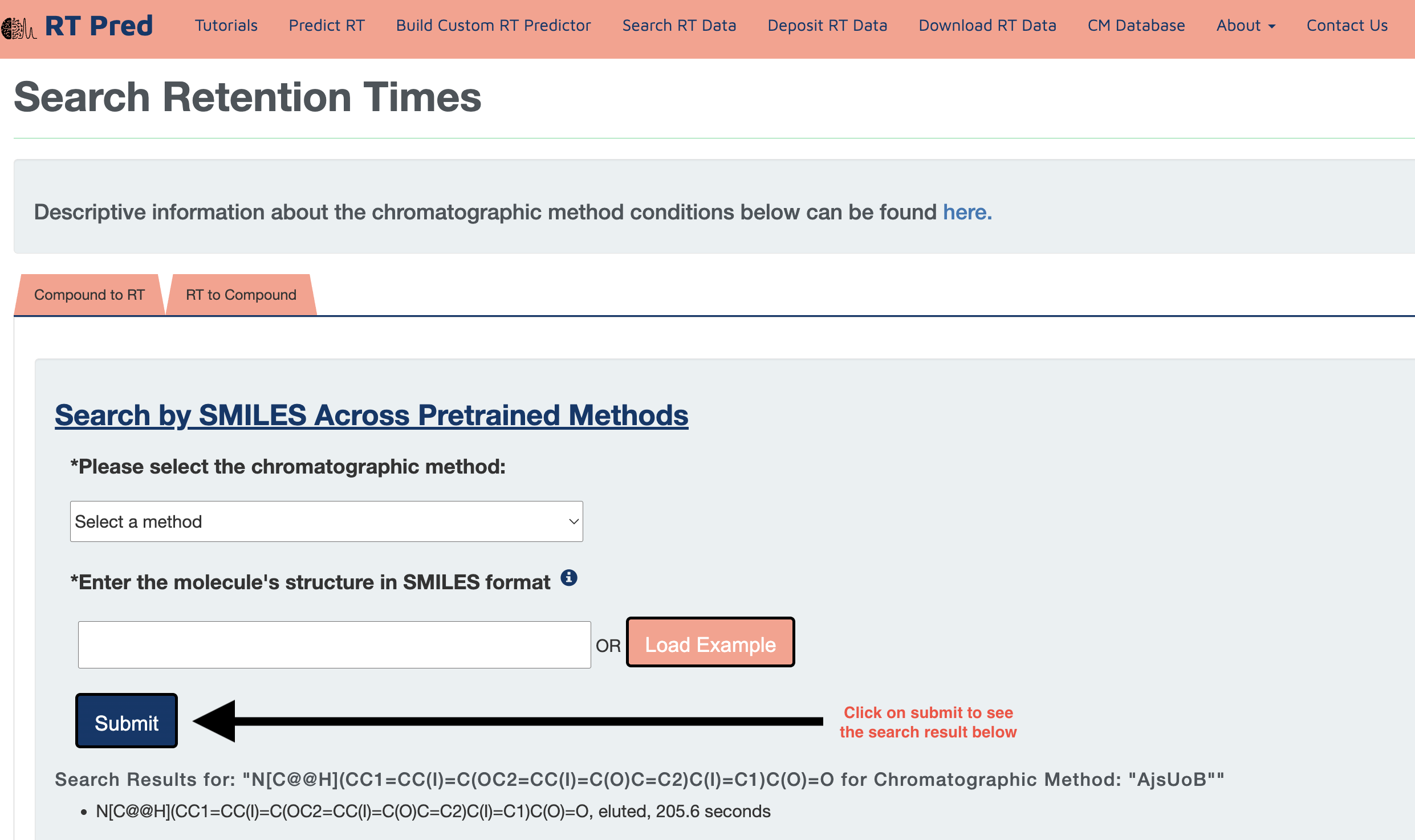
Search using RT to Compound
Step 1: Click on the "RT to Compound" tab to go the search module where compounds can be searched within a given range in a selected CM.
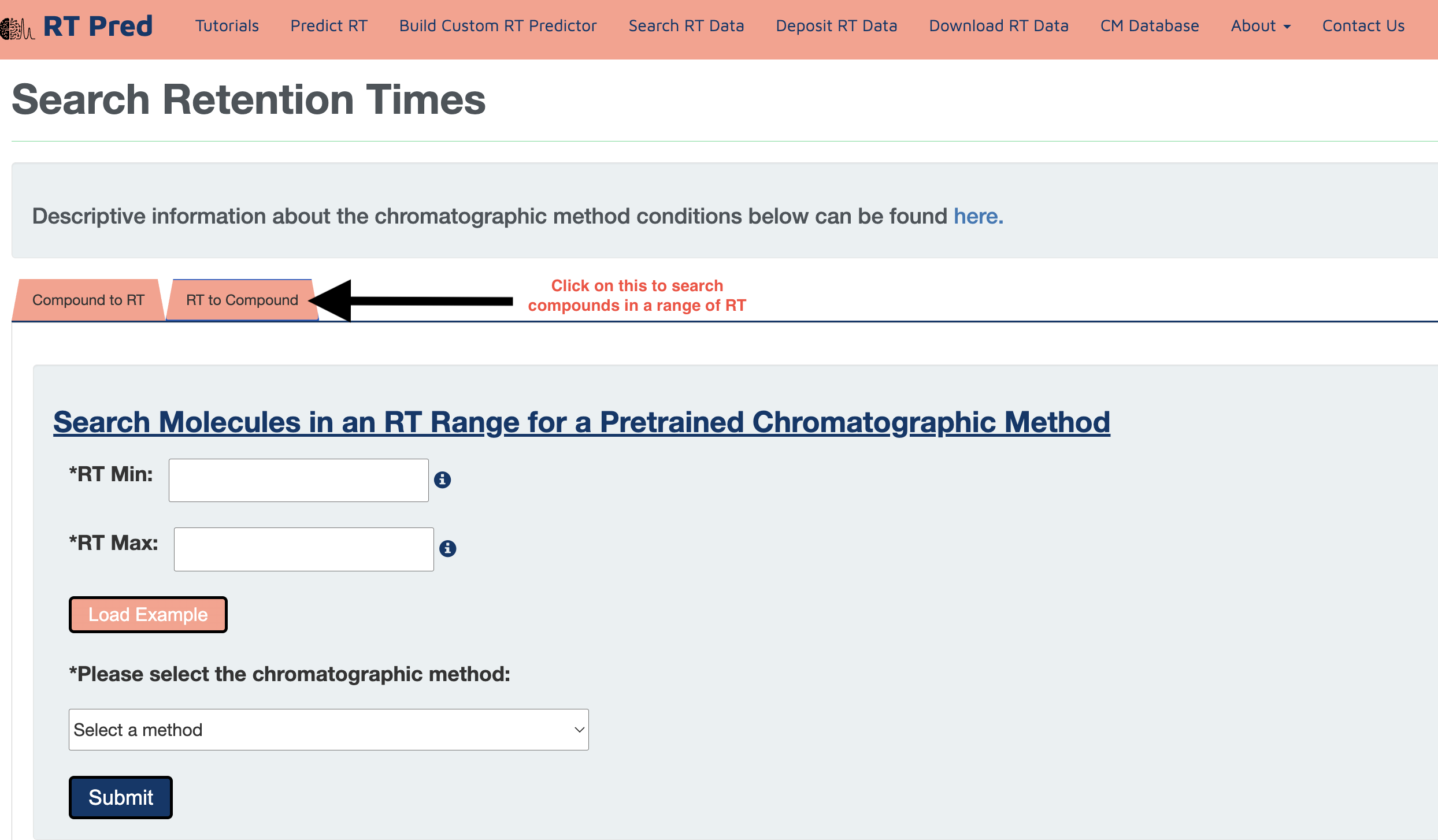
Step 2: Fill the boxes with two values, one should be the minimum value of the range and the other should be the maximum.
Alternatively, click on the load example button to add an example RT range.
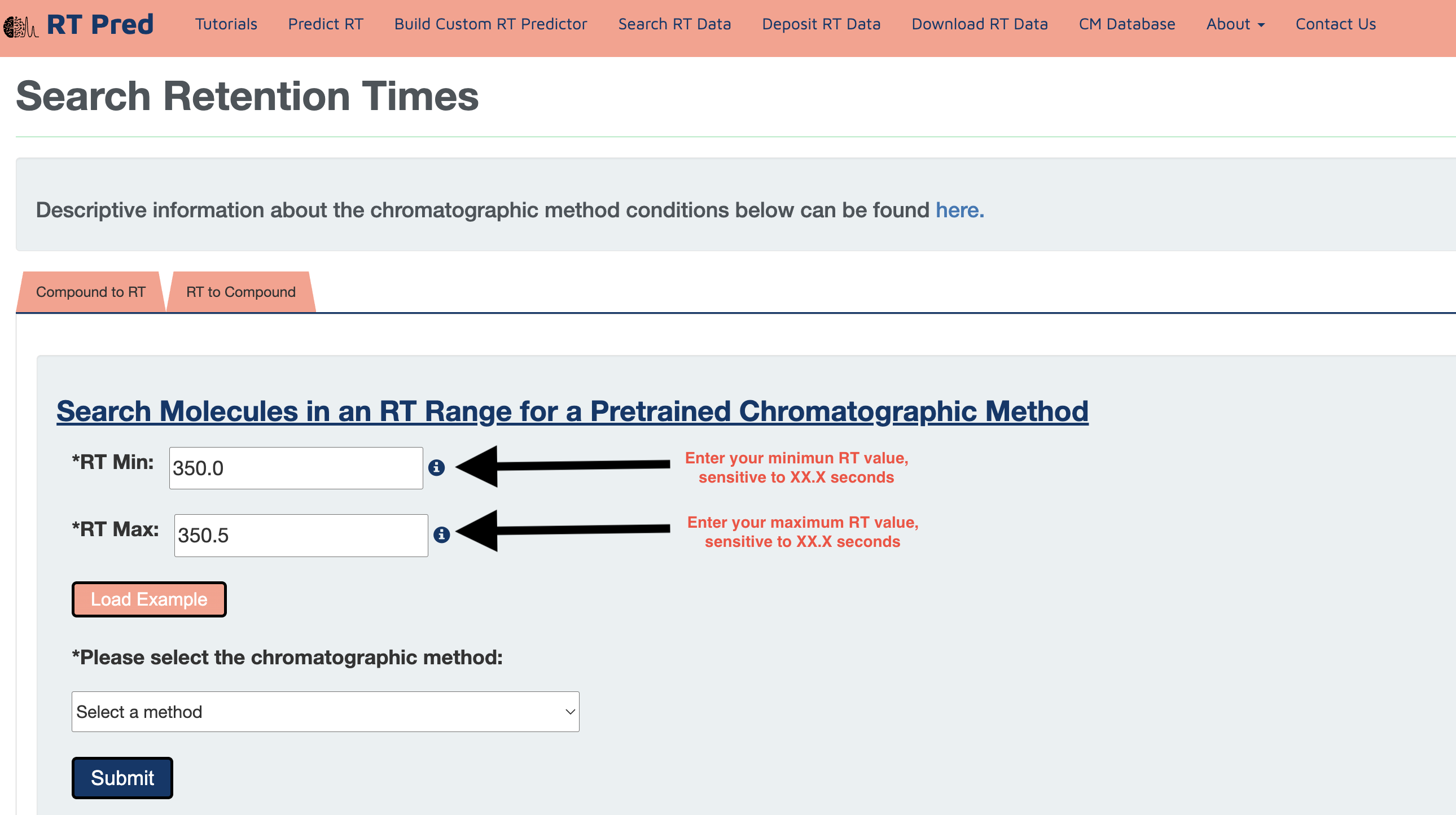
Step 3: Choose a pretrained Chromatographic Method from the drop down menu..
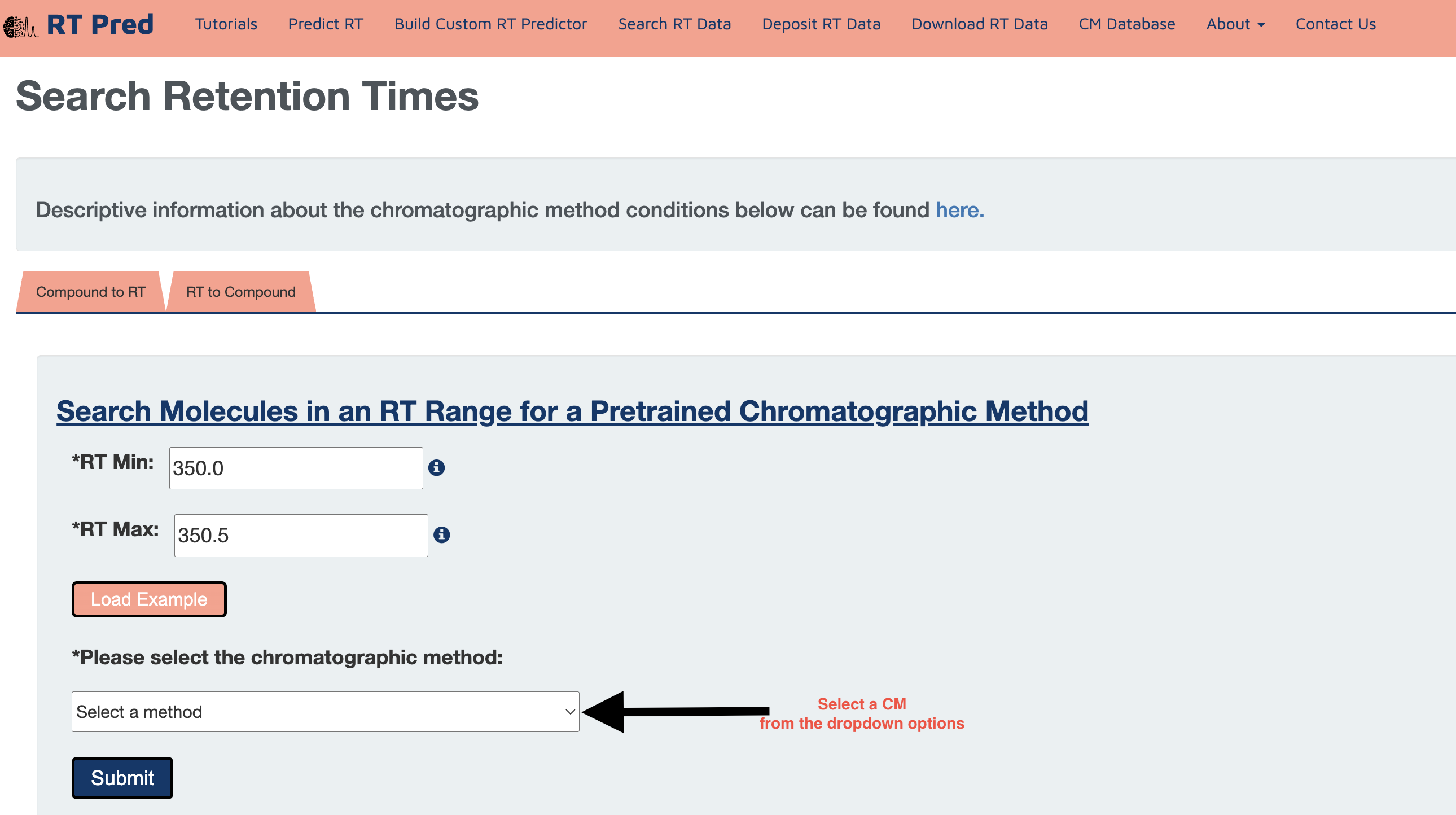
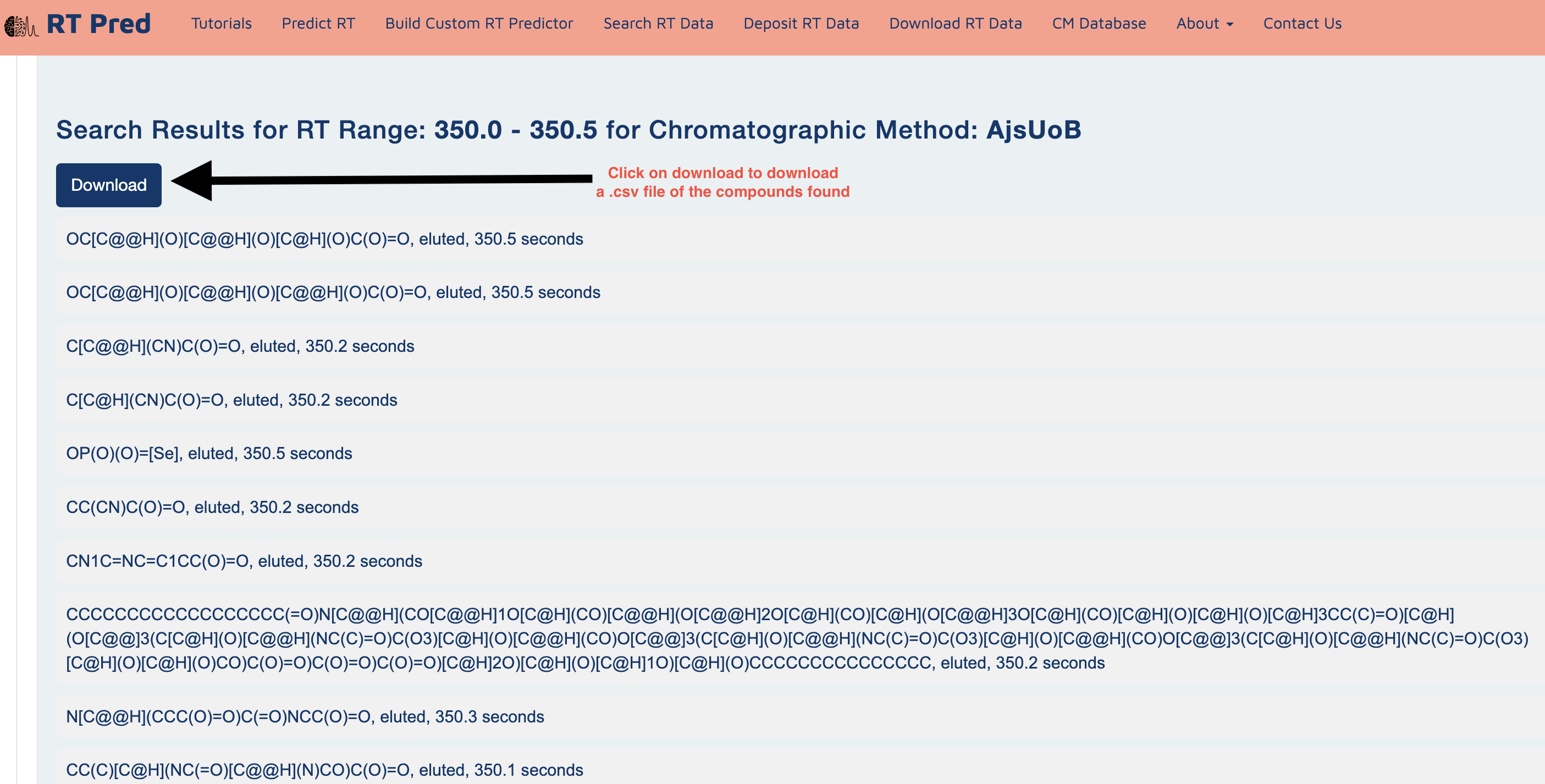
Step 4: Click on the submit button to search for the compounds with RTs within the given range in the selected CM.
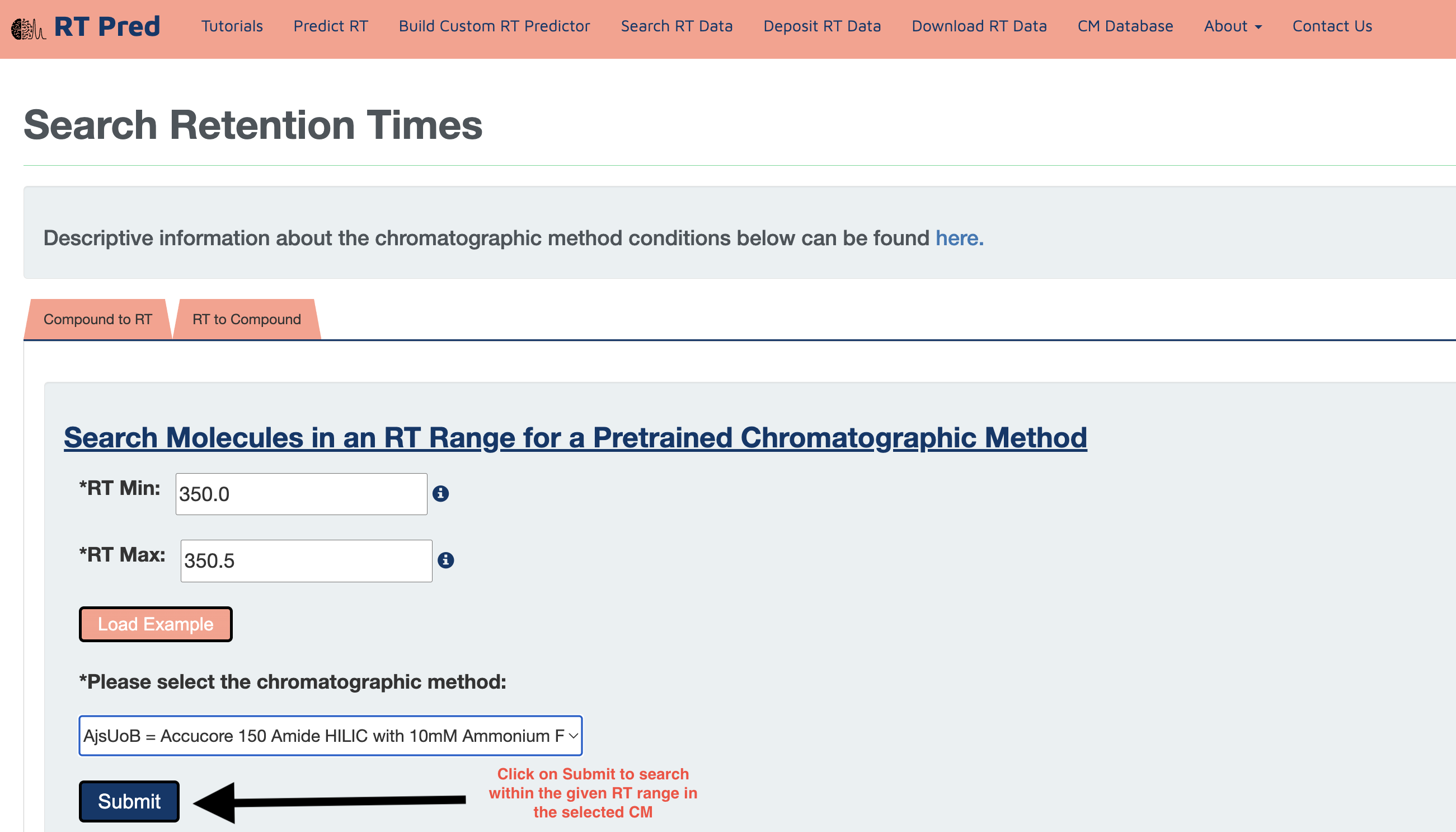
Step 5: Click on the Download button to download a csv file containing your results.